The mutations that drive cancer formation are often found in “hub” genes that regulate many aspects of cell growth and survival. But these key genes are not always good therapeutic targets — some are even considered “undruggable.” In the latest issue of GENETICS, Bailey et al. identify a strategy for fighting cancer cells that carry a mutation in one such difficult-to-target hub gene, FBW7. This tumor suppressor is mutated in approximately 6% of all cancer cases, including around a third of cases of T-cell acute lymphocytic leukemia and cholangiocarcinoma.
The FBW7 protein is part of the SCF ubiquitin ligase complex that marks other proteins for selective degradation. Many of these substrates are involved in oncogenesis (cancer formation), including the cell cycle regulator cyclin-E and the cell death inhibitor MCL1. Without FBW7 keeping them in check, these cancer-promoting proteins accumulate in the cell and can trigger uncontrolled division.
Even though FBW7’s tumor suppressor function is well understood, little is known about how the mutant cells might be targeted for treatment. That’s because inactivating the function of a tumor suppressor gene (e.g. with a drug) would encourage, not inhibit cancer, while many of FBW7’s oncogenic substrates are themselves difficult to manipulate with pharmaceuticals.
The authors approached this problem by screening for “synthetic lethal” partners of FBW7, which are proteins needed for cell survival when FBW7 is non-functional. They screened approximately 16,000 human genes by knocking down their expression in fbw7 mutant colorectal cancer cells and FBW7 wild-type cells, looking for proliferation effects that were specific to the mutants.
One of the candidates identified in the screen was BUBR1, which encodes a component of the mitotic spindle assembly checkpoint (SAC). This checkpoint prevents cell division from proceeding until all chromosomes are correctly attached to the spindle apparatus. When BUBR1 expression is knocked down in fbw7 mutant cells, the cells proliferate slower than wild-type and become more prone to losing and gaining chromosomes through cell division errors. This vulnerability of fbw7 cells was confirmed by tests with other SAC components.
Why do fbw7 cells have such a critical need for spindle assembly surveillance? One part of the answer is that the cells have dysregulated cyclin E. This is suggested by the fact that dampening expression of cyclin E rescues fbw7 mutant cells from their dependence on BUBR1. Cyclin E is not the whole story, however, because boosting levels of this protein alone was not enough to make FBW7 wild-type cells sensitive to SAC loss; this was only achieved by overexpressing another FBW7 substrate, MCL1, along with a form of cyclin E sometimes seen in cancer cells.
The results show that in this cell culture model, fbw7 mutant cells depend on the SAC for their cancerous potential, likely because they have cell cycle defects that necessitate extra time for spindle assembly. Without this breathing room, the dividing cells may be more prone to lethal chromosome segregation mistakes.
The authors suggest that other cancer cell types — many of which experience chronic chromosome instability — might also depend on the SAC. They suggest that exploiting this weak spot could be a promising strategy for developing anticancer drugs, especially since blocking SAC function could have fewer side-effects than existing anti-mitotic drugs. Though there is a long way to go before this idea could be applied in the clinic, the “undruggable” target FBW7 may have pointed the way to a more accessible chink in cancer’s armor.
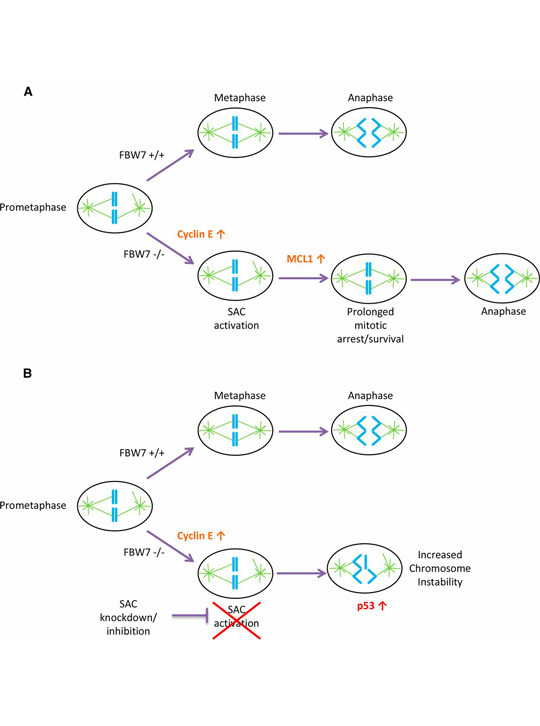
A model for SAC dependence in cells lacking FBW7. From Baikey et al. (A) FBW7 +/+ cells in prometaphase efficiently align their chromosomes and correctly segregate properly in anaphase with less reliance on the SAC. FBW7 −/− cells have an increase in cyclin E, which causes problems in mitosis and may lead to SAC activation. FBW7 −/− cells can survive prolonged SAC activation in part because of stabilization of MCL1 allowing more time for chromosome alignment. (B) A decrease in SAC activation by knockdown of BUBR1 may significantly shorten the time available for chromosome alignment in FBW7 −/− cells, resulting in improper chromosome segregation and intolerable levels of chromosome instability.
Citation:
Bailey, M. L., Singh, T., Mero, P., Moffat, J., & Hieter, P. (2015). Dependence of Human Colorectal Cells Lacking the FBW7 Tumor Suppressor on the Spindle Assembly Checkpoint. Genetics, 201(3), 885-895. Doi: 10.1534/genetics.115.180653































