Memory is a complex neurological process, and understanding its mechanics rarely yields clear-cut answers. So far, research on the memory-implicated gene HDAC4 has been no exception—in fact, HDAC4’s effects on long-term memory seem completely contradictory. Increased HDAC4 protein in the cell nucleus causes memory deficits, and consistent with this, a human mutation that resulted in HDAC4 being confined to the nucleus has been linked with brachydactyly mental retardation syndrome (BMDR) in at least one person. In mice, a similar HDAC4 variant causes cognitive deficits and reduced expression of genes associated with neural plasticity, which is essential for memory formation. But confusingly, loss of one copy of HDAC4 is also correlated with BDMR, and knockdown of HDAC4 in Drosophila melanogaster impairs memory. Research published in the July issue of GENETICS takes a step toward clearing the fog shrouding this protein’s strange behavior.
At first, the researchers wondered whether HDAC4’s effects on memory might be related to its role as a transcriptional repressor. But when they boosted levels of the protein in adult flies, they detected minimal changes in gene expression in the head, where transcriptional activity specific to memory would be expected to be found. This may be because changes in gene expression occur only in small regions of the brain and thus can’t be detected by examining the whole head. To probe more deeply, the researchers needed an alternative approach to detect interactions with HDAC4. For this, they turned to a unique assay.
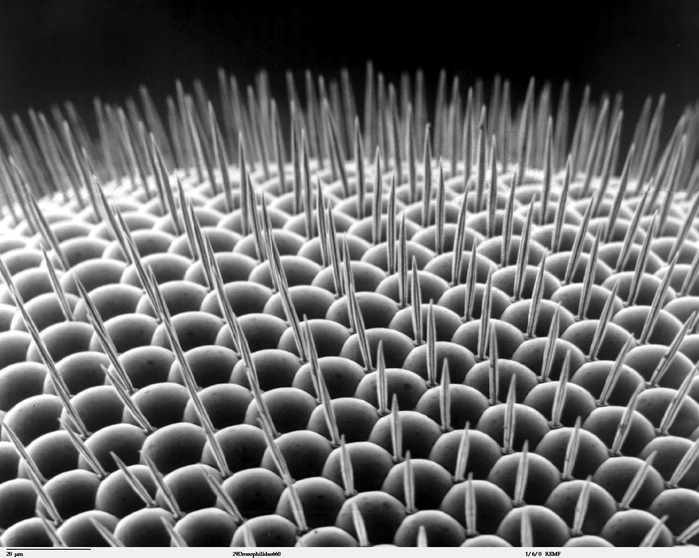
Previously, the authors had observed that overexpression of HDAC4 in the fly eye results in a “rough eye” phenotype characterized by disorganized ommatidia (optical units of the compound eye) and bristles. The severity of this disorganization increases with higher levels of HDAC4 overexpression. They used this assay as a readout for HDAC4 function when other genes in the fly were knocked down: if the targeted gene reduced or increased the severity of the rough eye phenotype, they concluded it interacted genetically with HDAC4.
The researchers focused their search on genes involved in synaptic plasticity, memory, neurological functioning, or chromatin modification, as well as genes known to interact with HDAC4 in other tissues or other organisms. They overexpressed HDAC4 in fly lines from the Vienna Drosophila Resource Centre that had RNAi knockdowns of the selected genes and looked for changes in the rough eye phenotype. From this screen, they found 26 genes that interacted strongly with HDAC4.
Probing further, they performed a functional network analysis using the Search Tool for the Retrieval of Interaction Genes (STRING) database, which predicts physical and functional links between proteins using data from experiments and predictions from computational studies. The genes clustered in a few major groups: transcription, the cytoskeleton, and SUMOylation. Interestingly, recent evidence has linked SUMOylation—a protein modification with many functions—to neuronal protein activity and memory formation. Because of this, the researchers decided to focus on Ubc9, one of the SUMOylation genes.
Follow-up experiments strongly suggested that Ubc9 and HDAC4 interacted genetically, but the researchers still needed to find out if this interaction was related to both genes’ effects on memory. To investigate this, they used a courtship assay, which measures a male fly’s ability to remember not to bother courting a female who has already rebuffed his previous advances. When the researchers knocked down Ubc9, the flies were less able to keep track of their courtship failures. Increasing levels of HDAC4 in the same flies produced a more severe derangement of long-term memory, providing evidence that Ubc9 and HDAC4’s memory-related functions are linked.
These results open up new research avenues for better understanding long-term memory, including the link between SUMOylation and HDAC4. Interestingly, some of the cytoskeletal genes that interacted genetically with HDAC4 are involved in axon and dendrite formation, implying that their roles may also be worth studying. Understanding the connections between HDAC4’s activity inside the nucleus and with the cytoskeleton and SUMOylation machinery could explain its seemingly conflicting roles in long-term memory—and, perhaps, help to illuminate some of the inner workings of memory in general.
CITATION:
Schwartz, S.; Truglio, M.;Scott, M.; Fitzsimons, H. Long-Term Memory in Drosophila Is Influenced by Histone Deacetylase HDAC4 Interacting with SUMO-Conjugating Enzyme Ubc9.
GENETICS, 203(3), 1249-1264.
DOI: 10.1534/genetics.115.183194
http://www.genetics.org/content/203/3/1249