With 1014 authors, an article by Leung et al. in the May issue of G3 has the largest author list of any paper published in the journal. More than 900 of those authors were undergraduate students when they performed the research.
Over several years, students at 63 higher education institutions across the US conducted an investigation far beyond the power of any individual lab. In so doing, this massive team not only advanced science, but gained valuable research skills and experience.
“By organizing the efforts of ‘massively parallel’ undergrads, we can solve problems that would defeat other methods,” says Genomics Education Partnership (GEP) program director Sarah Elgin. “At the same time, students learn how to handle the messiness of real data, to evaluate different kinds of evidence, and to justify their conclusions.”
The GEP is a large collaboration of college/university faculty coordinated from the Biology Department and The Genome Institute at Washington University in St. Louis. Their goals are to introduce bioinformatics into the undergraduate curriculum and to integrate research experience into the academic year. With this classroom-based approach, many more students can access educational opportunities normally restricted to a small number of summer research spots.
The project tackled by the GEP students was annotating and improving the sequence of the Muller F element of Drosophila fruit flies. The F element is so small that it looks like a compact “dot” alongside the other fruit fly chromosomes and is commonly called the “dot chromosome.”
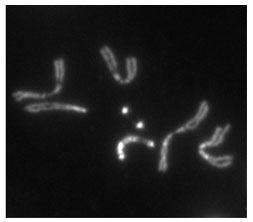
Heterochromatin staining in Drosophila melanogaster chromosomes. The two dot chromosomes are visible at center. Image source: flybase.org
Intriguingly, the dot chromosome is entirely heterochromatic by many criteria, meaning that it’s made up of tightly packaged DNA, a state typically associated with repression of gene expression, low recombination rates, and regions that contain few genes. But despite being packed into heterochromatin, the dot chromosome in Drosophila is not an inert gene desert; indeed, a region stretching nearly a third the length of the chromosome (the distal 1.3 Mb) has around the same density of active genes as expected in euchromatic (i.e., non-heterochromatic) genome regions.
How has this unusual chromatin context affected the evolution of the dot chromosome? To investigate this question, the GEP team wanted to compare the dot chromosome to a euchromatic region from a different chromosome. But this exploration required high quality assembly and annotated sequence from several different Drosophila species, not just D. melanogaster, the species in which the dot chromosome has been most intensively studied.
The students used publicly available draft genome sequences for three Drosophila species, plus the high quality sequence of D. melanogaster, species separated by 40 million years of evolution. In the first stage of work, they corrected errors, such as misassembly, in the published sequences and requested additional sequencing reactions to cover gaps. “The students do a significantly better job at improving the sequence than the software does,” says Elgin. “Most programs get bogged down in the repeat sequences that are abundant on the dot chromosome.”
The students then took the improved sequence and used multiple types of evidence to annotate the start, stop, and splice sites for each gene. The evidence used by the students included conservation, expression data, and computational prediction based on sequence features.
“We think a lot of the educational benefit of the project comes from asking students to weigh the evidence; sometimes it’s contradictory, sometimes one clue is more reliable than another, sometimes the students need to dig a bit deeper,” says Elgin. “Basically we’re teaching them to look carefully at data and be suspicious, be skeptical.”
Each chunk of annotation was completed by at least two independent groups of students, allowing them to cross-check their findings and fix errors.
The end result was a high quality data set that allowed the team, led by GEP staff member Wilson Leung, to statistically compare the properties of the dot chromosome to the euchromatic comparison region in all four species.
This comparison revealed that most of the distinctive properties of the D. melanogaster dot chromosome are conserved across species — including genes with longer introns and more coding exons on average than the euchromatic comparison region, as well as a higher density of repeat sequences. The accumulated repeats — mostly remnants of now inactive transposable elements — can partly explain why dot chromosome genes have larger introns (the introns contain more repeats), though it doesn’t explain why the genes tend to have more coding exons.
Dot chromosome genes also showed less evidence for selection — in the form of codon bias — than the euchromatic comparison regions. This agrees with theoretical predictions that natural selection should be less effective where recombination rate is low, such as in heterochromatin. However, for D. grimshawi (a Hawaiian species that has been geographically isolated from the others), there is greater evidence for selection on dot chromosome codon bias, suggesting a higher rate of recombination than in the other species. D. grimshawi also has a lower transposon density on the dot chromosome, so the authors suggest that the density and types of transposons may affect the degree of local heterochromatin formation.
Unexpectedly, the authors found that the GC content (and therefore melting temperature) of both genes and their flanking regions is significantly lower in the dot chromosome than in the euchromatic comparison regions. Elgin says this is one of the findings from the project that she finds most fascinating, because she can’t yet explain it. “It drives me nuts!” she says. One possibility could be that the lower melting temperature enhances the transcription efficiency of genes trapped in a heterochromatic context. This might be one way that expression of dot chromosome genes is maintained at similar levels to genes from euchromatic contexts.
There are plenty more insights to be mined from the data, says Elgin. “At some point though we had to ask Wilson to stop analyzing data, because we had to start writing. One of our big goals was to publish a paper with the students as co-authors. We wanted them to be able to look themselves up on PubMed!”
Although the GEP faculty and staff wrote the article drafts, each of the 940 students listed as a co-authors had to read and approve the manuscript before submission. “Actually we got some important comments back from students,” says Elgin.
The GEP also measures the program’s educational performance. Research published last year in CBE-Life Sciences Education shows that not only do these students increase their knowledge of genes and genomes, they also report gains in their ability to analyze data and understand the research process that are similar to students who had performed a summer research project. Both types of learning gain — knowledge and understanding of research — were more striking when more class time was devoted to the project. Given enough time (on average, around 45 hours of class time), GEP student gains even exceeded those reported by students who had spent a summer in a research lab.
“Faculty are sometimes skeptical that this kind of project will work for their students. But the GEP includes a diverse range of schools serving different types of students, and the learning gains were similar across every category we tested. I believe any student can benefit,” says Elgin.
More students are benefiting as the GEP expands and takes on new research projects. Some of these students will continue in research careers — as Wilson Leung did after his own undergraduate research experience in a precursor to the GEP. Many other students will choose different paths, but will do so with a richer understanding of both genetics and the hard work of building new knowledge.
Citation: Leung, W. et al. (2015). Drosophila Muller F Elements Maintain a Distinct Set of Genomic Properties Over 40 Million Years of Evolution. G3: Genes| Genomes| Genetics 5(5):719-740 doi: 10.1534/g3.114.015966 http://www.g3journal.org/content/5/5/719.full