When an eastern diamondback rattlesnake bites its prey, it injects a cocktail of toxic proteins and peptides that attack on multiple fronts. These toxins destroy blood vessels, block the blood clotting cascade, cause necrosis, and inflict crippling pain.
But the precise recipe for this noxious mix is generally thought to depend on where the snake was born. Cycles of co-evolution between predator and prey are expected to drive extensive genetic divergence between locations; the data so far have mostly fit this model, and venom composition seems to vary with geography.
In a recent issue of GENETICS, Darin Rokyta and colleagues reported the results of a large survey of venom diversity across two snake species sharing nearly identical ranges and similar habitats in the southeastern United States. As expected, the mix in one species—the eastern diamondback rattlesnake—varied considerably from place to place. But the eastern coral snakes told a completely different story. In contrast to its rattlesnake neighbors, no matter where a coral snake came from, its venom was always the same.
Rokyta says the team was shocked by this lack of variation. “This is the first time anyone has looked at venom variation at this scale, and everybody has assumed that the co-evolutionary arms race would cause local populations to diverge quickly.”
The results not only challenge this assumption, they provide crucial information for rattlesnake conservation and coral snake antivenom development.

When an eastern diamondback rattlesnake bites its prey, it injects a cocktail of toxic proteins and peptides that attack on multiple fronts. These toxins destroy blood vessels, block the blood clotting cascade, cause necrosis, and inflict crippling pain. But the precise recipe for this noxious mix is generally thought to depend on where the snake was born. Cycles of co-evolution between predator and prey are expected to drive extensive genetic divergence between locations; the data so far have mostly fit this model and venom composition seems to vary with geography. In a recent issue of GENETICS, Darin Rokyta and colleagues reported the results of a large survey of venom diversity across two snake species sharing nearly identical ranges and similar habitats in the southeastern United States. As expected, the mix in one species—the eastern diamondback rattlesnake—varied considerably from place to place. But the eastern coral snakes told a completely different story. In contrast to its rattlesnake neighbors, no matter where a coral snake came from, its venom was always the same. Rokyta says the team was shocked by this lack of variation. “This is the first time anyone has looked at venom variation at this scale, and everybody has assumed that the co-evolutionary arms race would cause local populations to diverge quickly.” The results not only challenge this assumption, they provide crucial information for rattlesnake conservation, and coral snake antivenom development. Image credit: Joseph Pfaller
Rokyta (Florida State University) and his colleagues study venom to generate powerful datasets for testing quantitative models of adaptation. These theoretical models of adaptation need to be evaluated and guided by data from the field, but the types of real-world traits evolutionary biologists typically study can’t be used in this way because they’re hard to link to specific genetic variants. That’s because these complex traits—like coat color, jaw shape, stress tolerance—are affected by many networks of molecular and developmental pathways, and many candidate genes are likely to contribute to their variation.
For venom, in contrast, the link between genotype and adaptive phenotype is relatively direct. Simple changes in expression of a particular toxin can boost or dampen venom potency, or alter its specificity for certain types of prey. And because venom is produced by a specialized gland, the genes encoding the toxins can be rapidly identified by analyzing the venom gland transcriptome. These data could help in the identification of relevant gene variants and the signatures of the corresponding proteins in proteomic analyses of venom samples. The bottom line? Rokyta sees analysis of snake venom as a shortcut to a detailed genotype-phenotype map of an ecologically critical trait and an ideal source of hard data to test and shape quantitative models.
Creating this genotype-phenotype map is a long term project, but as an important first step, Rokyta and his team have assembled the venom gland transcriptomes of the two snake species and collected venom and blood samples from individuals collected at many locations.
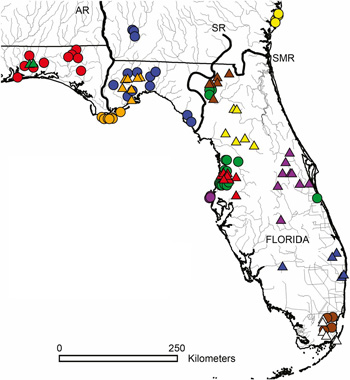
Venom samples were collected from 65 adult eastern diamondback rattlesnakes (circles) and 49 adult eastern coral snakes (triangles). Different colors represent different putative populations. AR, Apalachicola River; SMR, Saint Mary’s River; SR, Suwannee River. From Margres et al.
Using sensitive proteomic analyses, Rokyta et al. were able to divide the eastern diamondback samples into five different geographic groups that each brews its own brand of local venom. For example, in venom from snakes on Caladesi Island, near Tampa, Florida, two major toxins normally produced in this species are undetectable.
But for the coral snakes, there was no significant variation. Whether the snake had been caught in the Everglades or several hundred miles north in Florida’s panhandle, the venom was indistinguishable.
Rokyta says there are several possible explanations for this surprising result. One hypothesis is that differences in diet between the species might result in distinctive patterns of co-evolution. “Coral snakes tend to eat long, slender reptiles, while rattlesnakes tend to eat mammals,” he says. “So there may be big differences in the dynamics and genetics of their prey.”
Another idea is that the coral snake population could be less genetically diverse than the eastern diamondbacks, from a recent range expansion, for example, or a selective sweep. To figure out what’s really going on, the group is now examining the population genetics of the two species. “We have an intriguing pattern, but now we need to understand which driving selective forces—if any—caused this difference between the species.”
In the meantime, the results have practical significance. For example, they could aid efforts to conserve eastern diamondback populations. The species is currently in decline and under consideration for listing as threatened under the Endangered Species Act. These declines are thought to have been caused by habitat loss, compounded by persecution from humans. “We find a surprisingly large number of rattlesnakes dead on the road where people have obviously stopped, chopped the head and tail off, and left them for dead,” says Rokyta. He suggests conservation managers could use the new data to ensure the full range of local venom brews is conserved.
“Usually when you’re deciding which populations to focus your efforts on, you look at genetic differentiation,” says Rokyta. “But this type of functional differentiation in an adaptive trait might be even more useful; if you lose one of these five populations, that entire venom phenotype is gone.”
The lack of variation in eastern coral snake venom could be a boon for those developing a sorely-needed antivenom. Co-author Jack Facente (Agritoxins) runs a venom production lab to supply companies working on antivenom, and many of the coral snakes sampled in the study were originally collected by Facente for his lab. But if snakes born in different locations delivered significantly different venom, the resulting antivenom could also vary in effectiveness, depending on the range of subtypes sampled. Luckily for Facente, it doesn’t seem to matter where he finds his coral snakes—they always provide the same product.
Hopefully—after we learn more about the genetics of these beautiful snakes—we’ll one day know why.
CITATION:
Margres M.J., M. Seavy, K. P. Wray, J. Facente & D. R. Rokyta (2014). Contrasting Modes and Tempos of Venom Expression Evolution in Two Snake Species, Genetics, 199 (1) 165-176. DOI: http://dx.doi.org/10.1534/genetics.114.172437 http://www.genetics.org/content/199/1/165.full
































