Today’s guest post was contributed by Caitlan Rossi, a scientific and medical writer. Her work can be found at caitlanrossi.com.
Ever feel like you’re working for peanuts? When it comes to world food sustainability, geneticists argue, peanuts are the whole point.
Research published by Conde et al. in the latest issue of G3: Genes|Genomes|Genetics underscores the importance of peanut farming in the pursuit of global food security–and describes a new tool to make the legume even more resilient to climate change.
The study churns around Arachis hypogaea L.—better known as peanut or groundnut—which is commonly grown on small farms in developing countries for food, fodder, and profit. Over time, peanuts lost some of their resiliency as today’s allotetraploid plant was hybridized, domesticated, and adapted to meet consumers’ demands. In favor of higher yield and bigger, tastier nuts for consumers, breeders lost some of the genetic diversity that allowed the plant to fight certain diseases or withstand drought. Now, restoring genetic diversity is the key to developing better peanut breeding, and by extension, striving for global food security.
It’s a tough nut to crack, but that’s where core collections come in. These collections are groups of crop genotypes that reveal valuable molecular information, helping breeders map traits of interest. Core collections have enhanced the production of other crops like rice and wheat, and while several have been created for peanuts, they were developed before the advent of sophisticated genotyping technologies.
As groundnut breeders from various countries in Western Africa joined together with colleagues in Eastern and Southern Africa, the Groundnut Improvement Network for Africa (GINA) was launched. This collaborative group of ten breeding programs compiled more than 1,000 peanut varieties from nine countries—breeders were asked to nominate their preferred peanut lines—and analyzed them to create a core collection. In this new study, researchers explore the genetic diversity in those lines.
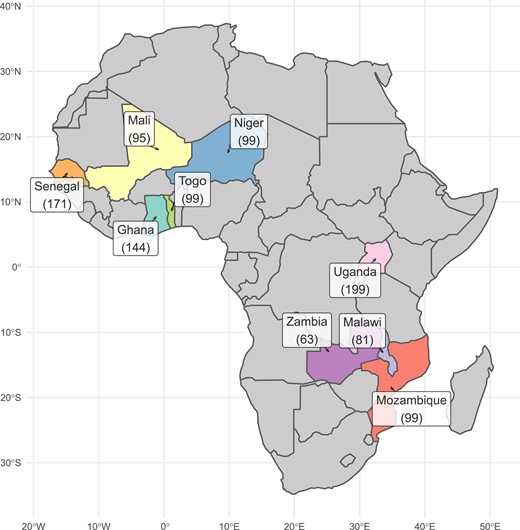
Using expected heterozygosity (He), a common measure for genetic variation within populations, the authors clustered groundnuts into three market types: Spanish, Valencia, and Virginia, and categorized them into one of two subspecies–most were fastigiata, while the rest were hypogaea.
When they dug deeper to analyze the genetic structure of the collection, the authors found that most of the breeding programs were rooted in an admixed genetic heritage, meaning they were a mix of multiple market types. This was especially true of the Valencia groundnuts. The authors also found that several lines of nuts had been nominated by more than one breeding program, suggesting past germplasm exchange. Some lines went by the same common name in different countries but were genetically different, demonstrating how difficult it can be to accurately record and manage germplasm.
The researchers developed a core collection of 300 accessions based on traits (breeder choices) and diversity (genetic distance-based sampling). While geneticists use this information to map favorable crop traits like resistance to disease, several peanut lines have already made their way into advanced release pipelines, spreading the agronomic and nutritional assets of this core collection to farmers and consumers.
The power of the collection lies in its collective genetic variability, a unique compilation of the many small but diverse collections throughout Africa. In a nutshell, this much-needed resource will help mobilize peanut diversity to make the plant—and the farmers that grow it—more resilient in the face of future challenges.
References
The groundnut improvement network for Africa (GINA) germplasm collection: a unique genetic resource for breeding and gene discovery
Soukeye Conde, Jean-François Rami, David K Okello, Aissatou Sambou, Amade Muitia, Richard Oteng-Frimpong, Lutangu Makweti, Dramane Sako, Issa Faye, Justus Chintu, Adama M Coulibaly, Amos Miningou, James Y Asibuo, Moumouni Konate, Essohouna M Banla, Maguette Seye, Yvette R Djiboune, Hodo-Abalo Tossim, Samba N Sylla, David Hoisington, Josh Clevenger, Ye Chu, Shyam Tallury, Peggy Ozias-Akins, Daniel Fonceka
G3: Genes|Genomes|Genetics. January 2024.
DOI: 10.1093/g3journal/jkad244




