As ancient humans spread across the globe from their evolutionary birthplace in Africa, they tended to lose a little genetic diversity at each step along the way. New settlements were probably often founded by small groups that carried only a subset of the total diversity present in their homelands. Successive rounds of this “founder effect” mean that today, modern indigenous populations living further from Africa along the old migration routes now have lower genetic diversity than those closer to Africa.
These marks of our prehistoric movements can still affect people and society today. Differences in genetic diversity between human populations can complicate certain social disparities, argue Stanford geneticists Noah Rosenberg and Jonathan Kang in a review published in the latest issue of GENETICS. But they find no evidence that genetic diversity is related to geographic patterns of economic development, contrary to a controversial finding reported by economists. In fact, Rosenberg and Kang’s reanalysis suggests the economists’ result was just a statistical fluke.
This finding doesn’t mean we should ignore genetic effects in societal processes, say the authors. They describe three established examples of interactions between genetic diversity and societal disparities: in forensics, bone marrow transplantation, and genomic studies of health. As an increasing number of economic studies investigate population-genetic variables, the authors emphasize that geneticists and economists need to draw a careful distinction between approaches based on genetic principles and those that treat genetic data in the same way as non-biological data—as just one among many possible variables.
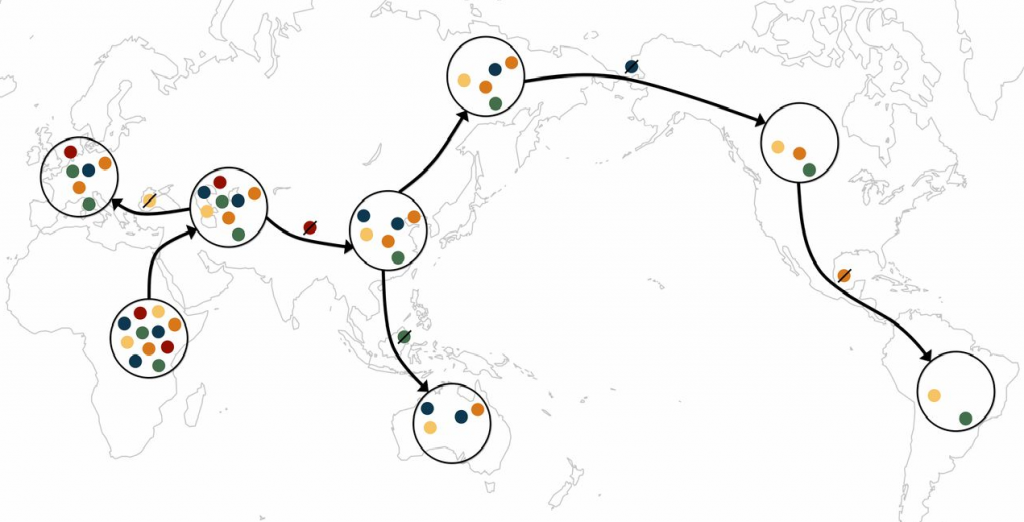
A schematic of the serial founder model. Each color represents a distinct allele. Migration events outward from Africa tend to carry with them only a subset of the genetic diversity from the source population, as some alleles are lost during migration events.
In 2012, Rosenberg was exploring the real-world impacts of differences in genetic diversity when an American Economic Review paper-in-press made a splash. The paper, by Quamrul Ashraf (Williams College) and Oded Galor (Brown University), argued that the intermediate levels of genetic diversity seen in Asian and European populations were optimal for their economic development, while low genetic diversity in Native American populations and high diversity in African populations had held their economic development back.
The paper was met with strong criticism from several prominent geneticists and anthropologists. A group of scholars from Harvard and the Broad Institute swiftly published an open letter criticizing the methods used in the project, claiming “Such haphazard methods and erroneous assumptions of statistical independence could equally find a genetic cause for the use of chopsticks.”
They also questioned whether pursuing such a project was ethical:
“….the suggestion that an ideal level of genetic variation could foster economic growth and could even be engineered has the potential to be misused with frightening consequences to justify indefensible practices such as ethnic cleansing or genocide.”
The debate grabbed Rosenberg’s attention. Like Ashraf and Galor, he was interested in how the genetic diversity differences between populations played out in societal processes. But the role of genetic diversity in the examples he was exploring was uncontroversial, in some cases even long established. What was the fundamental difference between Ashraf and Galor’s work and these examples?
In the process of examining this question, Rosenberg realized that he could make use of a much larger set of genetic diversity data that his lab had been preparing—237 populations from 39 countries subsequently published in the GSA journal G3: Genes|Genomes|Genetics, rather than the 53 populations in 21 countries studied by Ashraf and Galor. Using this expanded dataset and the economists’ own methods, Rosenberg and graduate student Kang repeated the analysis. They found no statistically significant relationship between genetic diversity and economic development when the larger dataset was used. They argue that Ashraf and Galor’s original result was likely a false positive, and that if they had happened to examine a different set of 21 countries, they would likely not have found a significant effect of genetic diversity in the first place.
In short, even if one disregards criticisms of the original study’s methods or ethics, it is likely the reported relationship between genetic diversity and economic variables was a coincidence.
But what about other real-world effects of genetic diversity?
Rosenberg and Kang’s first example is the use of familial identification in forensic genetic testing. Usually, when DNA is found at a crime scene, investigators look for a perfect match to a sample from law enforcement databases. But when no match can be found, it is becoming more common to search for partial matches that may identify relatives of the suspect.
The most famous example in the US is the 2010 identification and arrest of a suspect in the Los Angeles “Grim Sleeper” serial killer case. The suspect was tracked down through a partial match of crime scene samples to his son’s DNA profile from a database.
Critics point out that familial testing will intensify surveillance on those ethnic groups already overrepresented in law enforcement databases. The same groups will also bear more of the risk of being falsely implicated in a crime, a risk that is substantially higher for current familial identification than standard “perfect match” testing.
The population genetic features of different populations introduce another disparity to familial identification. Rori Rohlfs (University of California, Berkeley) and colleagues have shown that familial identification has a higher false positive rate in populations with lower genetic diversity, such as Native American groups.
Another example of the societal implications of genetic diversity is bone marrow transplantation matching. To reduce the risk of dangerous immune responses to a transplant, donors are chosen according to their genetic similarity to the recipient at six genes of the human leukocyte antigen (HLA) system. But for populations with higher genetic diversity, recipients have a lower chance of finding a match.
This genetic disparity can exacerbate the uneven availability of bone marrow transplants. Social factors that influence the likelihood someone from a particular ethnic group finds a match include the size of the population and their rate of inclusion in donor databases, and the rate at which members of the group participate as donors when they are identified as potential matches. African Americans seeking a donor have their chances of success lowered both because they are underrepresented in donor databases and because they belong to a population with higher genetic diversity.
The authors’ third example is the representation of different populations in genome-wide association studies (GWAS). GWAS have so far identified thousands of genetic links to hundreds of human traits and diseases. But by a 2011 estimate, 96% of GWAS participants had European ancestry. This extreme disparity means other ethnic groups are less likely to benefit from the results.
The reasons for the skew are partly sociological — the distribution of research funding, the structure of scientific collaboration networks, access to participants from each population — but such factors are exacerbated by genetic characteristics that make some populations more difficult to study by GWAS.
That’s because GWAS relies on the tendency for chromosomes to be passed on in chunks, co-inherited regions that show linkage disequilibrium (a statistical association between the alleles present at different loci). As a result, GWAS do not need to examine every possible genetic variant in the genome, but instead track a subset of a few hundred thousands or a million variants that each act as markers or tags for other variants within the same linkage disequilibrium “chunk”. Because recombination breaks up these chunks into smaller and smaller regions over the generations, and populations farther from Africa had smaller starting population sizes with fewer distinct chunks to disassemble, the diversity of chunks within a population increases with distance from Africa. African populations today have comparatively low linkage disequilibrium, which can in turn can mean more markers are needed to successfully “tag” the genome than for other populations.
As GWAS methods have matured, this technical issue has become less important, but in the early days of GWAS it helped limit research on African populations. This initial underrepresentation persisted as the existing European-skewed research was used in the development of new technologies and genomic resources.
The broader point of these three examples, says Rosenberg, is that each case includes sociological differences that contribute to the disparity – differences like population size, representation in databases, and funding—but he emphasizes that there are also contributions that relate to genetic diversity. To fully address these disparities, it will be important to take into account both types of effect: sociological differences and genetic diversity differences.
So what is the distinction between these examples and studies like that of Ashraf and Galor? Rosenberg says the arguments of the former were built using the theoretical machinery of population genetics. In comparison, the economists’ work looked only for correlations between genetic and economic variables.
Understanding this difference is an important part of evaluating claims about genetics, says Rosenberg, especially as studies investigating correlations between economic and biological variables become increasingly common. “They’re not necessarily wrong,” says Rosenberg. “They just don’t draw any support from principles of genetics.”
CITATION:
Noah A. Rosenberg and Jonathan T.L. Kang (2015). Genetic Diversity and Socially Important Disparities. Genetics, 201(1), 1-12. doi: 10.1534/genetics.115.176750
http://www.genetics.org/content/201/1/1.full













