The December issue of GENETICS is out now! Check out the highlights below or the full Table of Contents here.
Ectopic centromere nucleation by CENP-A in fission yeast, pp. 1433–1446
Marlyn Gonzalez, Haijin He, Qianhua Dong, Siyu Sun, and Fei Li
The mechanisms protecting the cell against formation of ectopic centromeres (neocentromeres) are poorly understood. Gonzalez et al. show that overexpression of the centromere-specific histone H3 variant CENP-A in fission yeast results in the assembly of ectopic CENP-A chromatin with features of neocentromeres. Intriguingly, this assembly is suppressed by overexpression of histone H3 or H4. The N-terminal domain of CENP-A prevented ectopic CENP-A assembly via ubiquitin-dependent proteolysis.
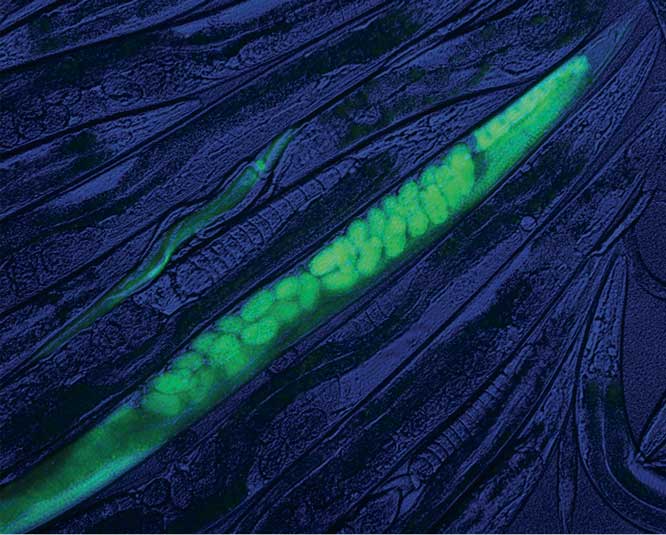
Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans, pp. 1347–1356
Alexandre Paix, Yuemeng Wang, Harold E. Smith, Chih-Yung S. Lee, Deepika Calidas, Tu Lu, Jarrett Smith, Helen Schmidt, Michael W. Krause, and Geraldine Seydoux
Paix et al. demonstrate that homology-dependent repair of Cas9-induced double-strand breaks is very robust in C. elegans and requires only short regions of homology. The practical implication is that it is possible to introduce edits of any size in the C. elegansgenome using oligonucleotides and PCR fragments with homology arms that are only 30-60 bases long.
The effects of demography and long-term selection on the accuracy of genomic prediction with sequence data, pp. 1671–1684
Iona M. MacLeod, Ben J. Hayes, and Michael E. Goddard
Dense SNP genotypes can be used to predict genetic or phenotypic outcomes in individuals for a complex trait, such as disease risk in humans and production traits in crops or livestock. MacLeod et al. evaluate the potential for improving the accuracy of genomic predictions by using sequence data compared to high density SNP arrays. They simulated data with contrasting population history, both with and without long-term purifying selection and tested two common analytical methods. The increase in accuracy achieved with sequence data varied considerably, demonstrating important dependencies and interactions with method of analysis, demographic history and long-term selection.
Transcriptome analysis indicates considerable divergence in alternative splicing between duplicated genes in Arabidopsis thaliana, pp. 1473–1481
David C. Tack, William R. Pitchers, and Keith L. Adams
Changes in alternative splicing of duplicated genes can generate new protein isoforms or alter gene expression. Tack et al. show that alternative splicing events have diverged considerably between duplicate genes in Arabidopsis. Most of the conserved splicing events showed divergence in event frequency, and most duplicated genes have diverged in alternative splicing-induced RNA decay.
A novel targeted learning method for quantitative trait loci mapping, pp. 1369–1376
Hui Wang, Zhongyang Zhang, Sherri Rose, and Mark van der Laan
Wang et al. present a semiparametric approach to QTL mapping that requires fewer restrictive assumptions about the data and accommodates machine learning algorithms with statistical inference. In simulations, the method was substantially less biased than other approaches, and it provided improved estimates and QTL rankings. When applied to a barley data set, the approach displayed less noise than composite interval mapping.
Bayesian inference of shared recombination hotspots between humans and chimpanzees, pp. 1621–1628
Ying Wang and Bruce Rannala
Several population genetic analyses have failed to detect recombination hotspots shared between humans and chimpanzees, despite their close genetic relationship. Using a Bayesian method and focusing on chromosome 21, Wang and Rannala analyzed chimp and human SNP data from recent resequencing projects. They also reanalyzed data from a previous study of human recombination hotspots in the MHC region. In both cases, the authors find strong evidence for shared hotspots between the two species.
The distribution of pairwise genetic distances: a tool for investigating disease transmission, pp. 1395–1404
Colin J. Worby, Hsiao-Han Chang, William P. Hanage, and Marc Lipsitch
Pathogen genome sequencing is an increasingly common tool in disease outbreak investigations, but the relationship between genetic distance and epidemiological factors is not well understood. Worby et al. investigated the distribution of genetic distance between isolates sampled during an outbreak, along with the effect of within-host dynamics and transmission bottleneck size. The authors derived an approximation to this distribution for testing transmission route hypotheses. This approach avoids the intensive computation of many existing methods but can identify a greater proportion of transmission routes.
Epigenetic control of learning and memory in Drosophila by Tip60 HAT action, pp.1571–1586
Songjun Xu, Rona Wilf, Trisha Menon, Priyalakshmi Panikker, Jessica Sarthi, and Felice Elefant
Xu et al. demonstrate that the histone acetyltransferase (HAT) Tip60 plays an epigenetic transcriptional regulatory role in cognitive function in Drosophila. Dysregulation of Tip60 HAT levels in the mushroom body causes memory deficits and misexpression of Tip60 cognition-linked targets. Remarkably, the learning and memory deficits induced by Alzheimer’s disease-associated amyloid precursor protein (APP) were rescued by increasing mushroom body Tip60 HAT levels. These results highlight the clinical potential of HAT activators.