Today’s guest post was contributed by Maria Costanzo of Stanford University. She has been a biocurator since before the term was coined and has contributed to genome database projects for a variety of fungi. The views expressed are her own. Follow her on Twitter: @mariaccostanzo.
When someone asks what I do for a living, my answer is usually met with a blank stare. Biocuration is a rare and seemingly arcane occupation.
In many ways, curation of biological data is much like traditional curation of museum collections. Museum curators start with a collection of objects, and so do biocurators. While museum curators might select fossils or pottery shards, biocurators sift through facts—pieces of data—that are published in scientific journals.
Biocurators collect, organize, interpret, and display biological data, a task that is becoming more and more necessary. Genome sequences, protein functions, mutant phenotypes, gene expression data, and much more are published every day. It’s an impossible task for any individual scientist to keep up with so many papers. Biocuration makes it possible for researchers to access, synthesize, and analyze all these different types of information. It’s slowly becoming recognized as an occupation, and biocurators now have their own professional organization, the International Society for Biocuration.
Although researchers know of biocurators’ existence (because they use the databases that we maintain), most are still unaware of what it takes to get their data into those databases. When I go to conferences to represent the Saccharomyces Genome Database (SGD), where I’ve worked for more than a decade, one of the most frequent questions I’m asked is “So, what do you actually DO?”
First, curators have to find papers that are relevant to our particular curation effort. It’s relatively easy to identify articles on budding yeast, because most authors who publish on S. cerevisiae mention the species. It also helps that S. cerevisiae gene nomenclature is uniform and adheres to standards agreed upon by the research community and mediated by SGD.
For biocurators working on species other than S. cerevisiae, even identifying the right papers to curate can be a tough task. Researchers publishing on mammalian genes, especially, have a tendency to mix and match species in their experiments, and sometimes they don’t clearly identify the species of origin of the genes they study. Plus, gene naming is not nearly as uniform in some other species as it is in budding yeast. As an extreme example, the fruit fly Andorra gene is also known as “and”—try searching for papers about the and gene!
Once they have a set of relevant papers in hand, biocurators must decide which facts to select from those papers, just as museum curators select representative artifacts to display. The type of facts differs somewhat between databases: while some basic information, such as protein function or localization, is relevant to any organism, other biological phenomena, such as alternative mRNA splicing or regulatory microRNAs, are known only in certain species.
Biocurators need to record each fact so that it is traceable both to specific entities (e.g., a gene or protein, with a specific sequence, from a specific strain of a specific organism) and to the fact’s source (the publication). The way in which the fact is recorded matters too. Rather than just jotting down notes, biocurators have created “controlled vocabularies” that use specific phrases to express biological facts across genomes and even across organisms. The most well-known controlled vocabulary is the Gene Ontology (GO), which is used to express the molecular functions, biological roles, and subcellular locations of gene products from dozens of different species.
Next, biocurators face yet another decision: how to present the information on the web pages of the database. Museum curators engage visitors by designing visually attractive, easily understandable displays, and it’s not too different for biocurators. We’re helped in this effort by talented programmers and web designers, but a lot of scientific judgment goes into planning how to organize and display biological information so that it’s easy for scientists to search for and retrieve the results they need.
Now the analogy to museum curators breaks down, as the other parts of a biocurator’s job don’t have direct equivalents in the museum. Because visitors to a biological database are often looking for very specific pieces of information—about a gene, a pathway, a part of the cell—having efficient search tools is critical. Again, both programming and scientific expertise goes into designing these searches.
Finally, while museums employ guards to make sure that visitors don’t take anything with them when they leave, an important part of a biocurator’s job is to make it easy for visitors to take away large quantities of information. Research scientists need to be able to download data. So biocurators need to make sure to collect significant datasets and make them easily accessible, and also to load data into versatile data warehouses that allow custom queries, such as the InterMine software. SGD’s instance of InterMine, called YeastMine, contains virtually all of the S. cerevisiae data found in SGD and allows researchers to slice and dice it in countless ways. Being able to play with the existing data helps them to develop the hypotheses that will drive the next series of experiments, generating more data that will be published and incorporated into databases (see figure below).
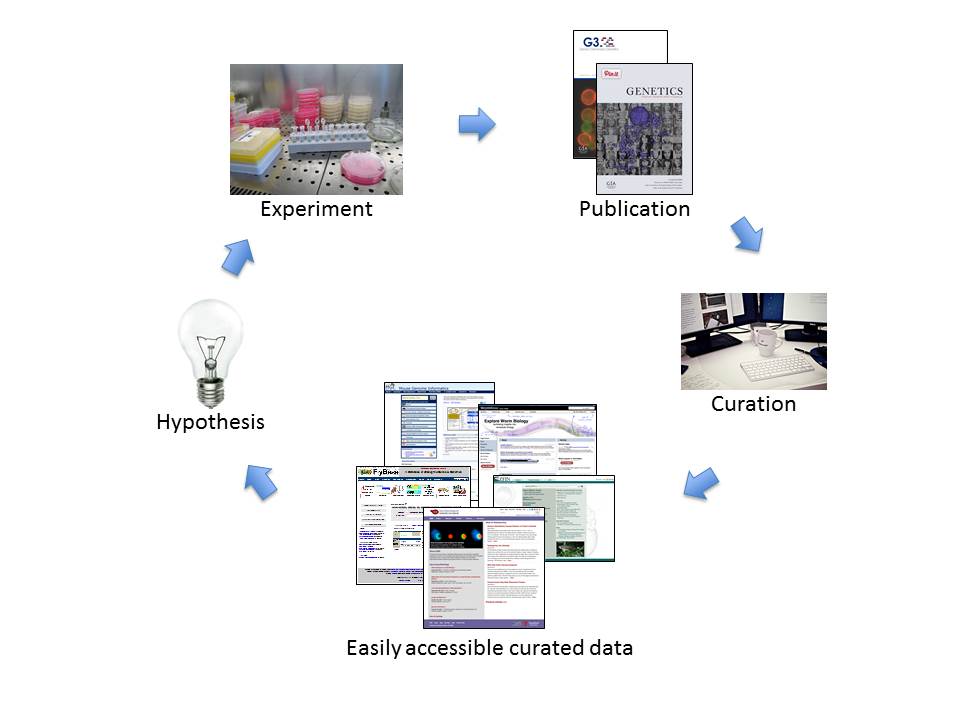
Researchers form hypotheses, design experiments to test those hypotheses, and publish the results. Biocurators then extract defined types of data from the publications, organize the information, and make it easily accessible in online databases. Using the databases, researchers can analyze, browse, and download the curated data, generating insights that allow them to formulate new hypotheses and start the cycle again. (Image Credit: Maria Costanzo)
Circling back to the original question, the simple answer to what biocurators actually DO, is this: They read a lot. Think a lot. Draw on their PhD-level scientific training to understand and interpret experimental results. Track down missing pieces of information to solve puzzles. Put everything in its proper place.
At times, for example when navigating the finer details of ontology structure or when confronting a list of hundreds of genes whose annotations need to be reviewed, the work can feel esoteric and isolated—a monkish discipline, like hand-copying the pages of an illuminated manuscript. But most of the time, it is exhilarating to have a broad view of the newest discoveries in your field and to have the opportunity to facilitate research all over the world. And when you attend a conference, the grateful scientists who rely on and appreciate your database can make you feel like a celebrity. Biocuration is a great way for those of us who don’t want to spend our careers standing at a lab bench to use our expertise and contribute to scientific progress.
The top 5 things researchers can do to ensure their work is curated accurately 1. Clearly identify the organism and the principal genes/proteins in the title or abstract of your paper. The initial screen that biocurators use to find relevant literature often searches titles and abstracts but not the full text of papers. 2. Provide accession numbers, standard names, and/or references for all the biological entities in your publication: the organism, strain or sub-strain, gene or protein sequence. Use accepted, official gene nomenclature. Don’t re-name a gene in your paper without consulting nomenclature authorities for your organism. Is it really worth choosing your own new gene name if it means that no one can figure out which gene you’ve characterized, and if your work isn’t linked correctly in databases? 3. Provide supplementary data in a format accessible to biocurators who may be using the files to load data: tab-delimited text or Excel files rather than PDFs. Please use a format that allows for the data to be easily reused and manipulated. 4. Write clearly. Biocurators are experts in their fields—typically, PhD-level scientists—but they are not specialists in every sub-discipline. Clear writing with a minimum of jargon will benefit all readers of your work! 5. Stay in touch. When you use your favorite database, remember that there are people behind the curation. If you have new data coming down the pipeline, or if you see something amiss in the database, let biocurators know. They’ll be happy to hear from you! |
| The views expressed in guest posts are those of the author and are not necessarily endorsed by the Genetics Society of America or its employees. |