The most diverse of all human genes encode a set of proteins at the frontline of our immune system. Many different Human Leukocyte Antigen (HLA) proteins are encoded by genes clumped together in one portion of the human genome known as the major histocompatibility complex region. HLA proteins sit on the surface of cells and bind the chopped-up fragments of other proteins (antigens), presenting them for inspection by immune cells. If the presented antigens are recognized as foreign, the immune system may be triggered to attack, whether the invaders are pathogens, cancer cells, or transplanted tissue.
Remarkably, most HLA genes have dozens, or even hundreds of alleles present in the human population, so across the genome region as a whole there are thousands of different alleles. This variation can affect individual susceptibility to infectious and autoimmune diseases, and is of great interest to geneticists studying human evolution and population history.
But despite the functional and evolutionary importance of HLA genes, sequencing data from this region is biased in many population genomics studies. As a consequence, the results from this region are often treated as suspect, and in many cases are discarded from subsequent analyses.
The reason is that it’s difficult to make sense of HLA data generated by the next-generation sequencing (NGS) methods that are now standard for population genomics studies. NGS methods generate short sequence reads, and when these reads come from highly polymorphic genes like the HLA genes it can be challenging to correctly align them to the genome reference sequence. This problem is even worse when the gene is just one of a group of related polymorphic genes, as is the case for many of the HLA loci.

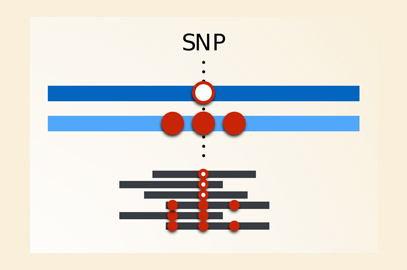
Genotyping errors for a highly polymorphic gene: The left hand side represents a case where sequence reads come from an individual who is heterozygous at a SNP, but where the rest of the gene is relatively similar to the reference for both haplotypes. The reads from both haplotypes can be aligned to the reference, and the SNP genotype is “called” (i.e. determined by the analysis software) correctly. The right hand side represents a case where one of the haplotypes is different to the reference sequence at more than one position. Reads from this haplotype won’t align with the reference and the genotype will be incorrectly called as homozygous at the SNP of interest. Image credit: Vitor R. C. Aguiar.
Though HLA loci are the worst-case scenario for this problem, other examples of polymorphic genes that come in related groups might suffer similar issues (such as the killer-like immunoglobulin receptor (KIR) and olfactory receptor genes). But because the degree of polymorphism in other gene families is less extreme than in the HLA genes, the analysis issues may be less obvious and therefore less likely to be accounted for.
In the latest issue of G3, Brandt et al. demonstrate the scale of the challenge using HLA data from the 1000 Genomes project, which is a collection of high-coverage exome and low-coverage whole-genome sequences from 1092 people generated by NGS. The authors compared the NGS data to a parallel dataset in which 930 of the samples from the 1000 Genomes project were re-sequenced using the “gold-standard” of Sanger sequencing, which doesn’t suffer from the same problems of short read alignment (the Sanger data were generated by Gourraud et al.)
Using the Sanger data as a benchmark, Brandt et al. showed that approximately 19% of single nucleotide polymorphism (SNP) genotypes for HLA genes in the NGS data were incorrect. And around a quarter of HLA SNPs had allele frequency estimates that differed between the two datasets by more than 0.1, with a bias towards overestimation of allele frequency in the NGS data. They also found that the most “unreliable” SNPs in NGS data were those with the highest heterozygosity. In other words, the SNPs at which people were mostly likely to be heterozygous were those that were most difficult to genotype correctly.
The results also suggest the NGS problem probably can’t be solved by boosting the intensity of sequencing efforts (i.e. increasing coverage). Rather, the authors’ argue that better computational analysis is the way forward. For example, they suggest that a major part of the problem is that standard approaches align reads to a single reference sequence. For HLA genes, and perhaps other polymorphic genes, alignment to a database of multiple reference sequences (for example, Boegel et al. and Dilthey et al.) can greatly improve genotyping accuracy by accounting for the different alleles possible at each gene.
A computational fix would be a boon to the many genetic studies that currently struggle to characterize HLA sequence data, including efforts to seek disease associations, quantify gene expression changes, and examine population histories. After all, the diversity of HLA genes is not only a technical challenge, but also a mark of their profound importance to immune system function and human survival.

Genotype mismatches between the 1000 Genomes (next-generation sequencing) and PAG2014 (Sanger sequencing) datasets. Results per polymorphic site (“Position”) and per individual. Dark squares indicate mismatches between genotypes in the two datasets. From Brandt et al.
CITATION:
Brandt, D.Y.C, Aguiar, V.R.C., Bitarello, B.D., Nunes, K., Goudet, J., & Meyer, D. (2015). Mapping Bias Overestimates Reference Allele Frequencies at the HLA Genes in the 1000 Genomes Project Phase I Data
G3: Genes|Genomes|Genetics, 5(5):931-941 doi: 10.1534/g3.114.015784
http://www.g3journal.org/content/5/5/931.full