Today’s guest author is Emma Farley, a postdoctoral researcher in Mike Levine’s lab, which recently relocated to Princeton (formerly at University of California, Berkeley).
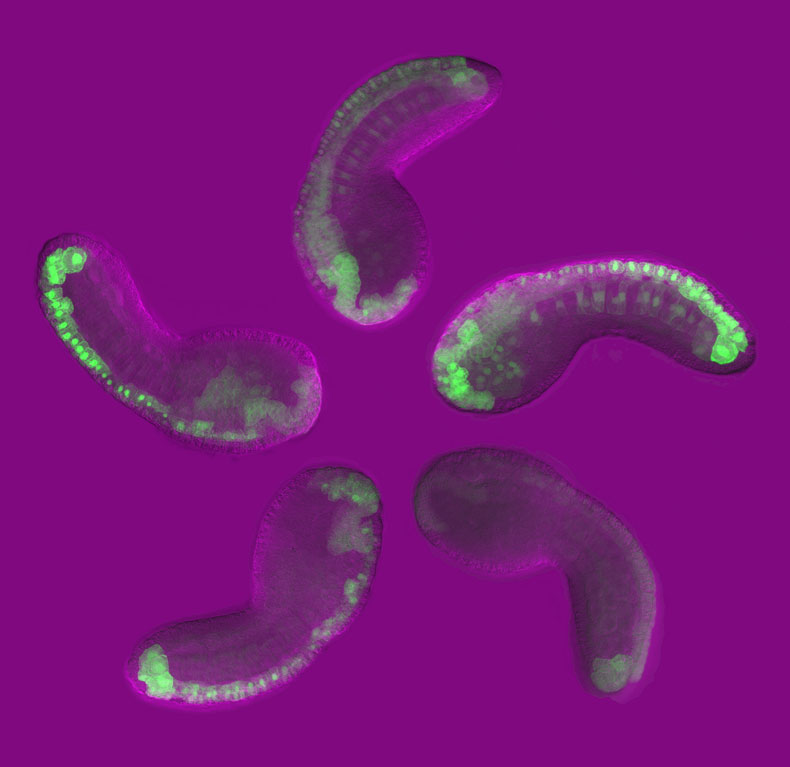
Sea squirts (Ciona intestanalis) are a classic system for the study of development. They were a favorite of early developmental biologists like Laurent Chabry, Ed Conklin, and Thomas Hunt Morgan. Over the course of 8 hours, you can watch a fertilized egg transform into a tadpole with a morphology indistinguishable from its vertebrate relatives! What’s more, the tadpole has only 2,500 cells. We can trace every single cell from the single celled fertilized egg to the fully formed tadpole because, unlike their vertebrate counterparts, sea squirts have defined cell lineages.
The sea squirt may have its roots in early developmental biology, but it’s even more relevant today. In fact, it’s uniquely suited to become a powerful tool for decoding the regulatory genome and in understanding the evolutionary origin of vertebrate-specific traits.The development of a sea squirt embryo follows a pattern similar to its vertebrate counterparts. This is not surprising when you consider that the sea squirt is a member of the ascidians, the sister group to the vertebrates. Sea squirts have been an ideal model organism for understanding the development and evolution of vertebrate-specific traits such as the origin of the two chambered heart and the origin of the vertebrate head. Another advantage of the Ciona is that its genome lacks the duplication seen in its vertebrate relatives, which makes it a simpler system for understanding development. For example, development of the Ciona heart uses the same genetic cues as mouse and humans, but is less complex, without the presence of multiple related proteins. These advantages all make Ciona an ideal system for studying the evolution of novel traits (e.g., neural crest and cranial placodes).
But there’s more to Ciona than evo-devo. As a postdoc in Mike Levine’s lab at UC Berkeley/Princeton, I was drawn to Ciona partly because we could introduce DNA constructs into thousands of embryos in a single hour-long experiment! Ciona—with its small number of cells, defined cell lineages, ease of imaging, and ease of introducing DNA—is an ideal system for high-throughput functional genomics, namely, how the genome encodes animal development.
The instructions for tissue-specific gene expression are located in enhancers, segments of DNA that act as switches to turn gene expression on and off at the right place and the right time. While we know that enhancers contain binding sites for particular transcription factors, that is only the first step in uncovering how enhancers control gene expression; this process tells us little about the types of mutations that can impact enhancer function.It’s apparent that humans need their heart-related genes turned on in the heart rather than, say, in the skin. So how does the sequence of an enhancer ensure genes are turned on only in the right location? Enhancers contain hundreds of nucleotides and multiple binding sites for transcription factors. This means that even the smallest enhancer has 1030 potential sequence combinations and several hundred thousand possible “grammatical variants” (i.e., differences in the order and orientation of binding sites)! To understand how changes in enhancer sequence impact where and when specific genes are turned on, we need high-throughput functional assays in developing embryos. They allow for an assay in every tissue type and also ensure the same conditions the genome naturally experiences.
Importantly, several hundred thousand Ciona embryos can be electroporated in a single experiment. This makes Ciona an ideal system in which to carry out high-throughput functional genomic assays to understand how enhancers instruct development. As a postdoc, I conceived and developed Synthetic Enhancer Library-Seq (SEL-Seq), a method to create millions of enhancer variants, and then tested these variants for function in developing Ciona embryos.
We used this technique to investigate how an enhancer important for brain development turns on gene expression. We identified the sequences within the enhancer that are necessary and sufficient to turn on expression in the brain. Surprisingly, we found that these sequences possess a number of suboptimal properties (affinity of binding sites and spacing of binding sites) that make them poor for activating gene expression. When we improved these features, the enhancer went “haywire,” leading to expression in other tissues. We proposed that suboptimal features are used so that enhancers avoid accidentally turning genes on in the wrong part of the body.
The genome is filled with multiple enhancers that activate expression of the same gene in the same place. Why are so many enhancers responsible for the same job? Clusters of weak enhancers such as “super-enhancers” could circumvent a trade-off in expression specificity and activity, and generate robust but localized patterns of expression. Because switching on a gene in the wrong tissue can lead to disease, understanding the principles governing enhancer specificity should help us identify mistakes in our DNA that result in problems.
While Ciona is a model organism that is often dismissed as a boutique system, a closer look reveals that the sea squirt is far more powerful than its name! It is the perfect system for deciphering the instructions for development embedded in the genome.

Emma Farley
Originally from Kent – the garden of England – Emma started in the Levine lab working on flies, but was quickly drawn to Ciona and the power of electroporation to decipher the regulatory code of the genome. Her specialty is transcriptional elongation and enhancer grammar. Ask her anything about enhancers – she enjoys discussing when, where, and why these strings of DNA sequence activate gene expression.
The views expressed in guest posts are those of the author and are not necessarily endorsed by the Genetics Society of America.


![Sea squirts. Image credit: Richard Ling [CC-BY-SA-2.0].](https://s36063.pcdn.co/wp-content/uploads/2015/11/8432568984_cca99cd469_z-300x225.jpg)
![Sea squirts. Image credit: Prilfish [CC-BY-SA-2.0], via Wikimedia Commons](https://s36063.pcdn.co/wp-content/uploads/2015/11/930351131_55de272264_z-e1447187140468-300x230.jpg)










