In 1979, as the US slid into recession, researchers began systematically crossing eight distinct inbred rat strains. Their goal was to establish a genetically diverse rat colony to serve as a base for phenotype measurements and artificial selection. But the creators of the NIH rat Heterogeneous Stocks (HS) faced major challenges:
“…the main one being that of supporting the colony until adequate demand arose to justify its existence. Maintaining an outbred colony without a clear-cut reason, given the current economic situation, is a difficult task.”
—Carl Hansen and Karen Spuhler, 1984. Development of the National Institutes of Health Genetically Heterogeneous Stock Rat. Alcoholism: Clinical and Experimental Research 8:477-479
Thirty years, many rat generations, and several recessions later, demand for the HS colony is rising for a clear-cut reason: the population is a unique resource for high-resolution quantitative trait loci (QTL) mapping in rats.

A group of Heterogeneous Stock rats show diverse phenotypes. Image credit: Katie Holl and Leah Solberg Woods.
Rats have long been workhorse models for physiology and pharmacology, but they are also becoming increasingly popular as models for disease genetics. The HS rats are a powerful population for mapping complex traits like disease because each individual’s genome is a fine-grained mosaic of the eight founder genomes. After more than sixty generations of crossing, their genomes have been recombined into much shorter chunks than in conventional two-parent crosses. That means QTLs can be mapped to much smaller regions, giving a more manageable list of causal genes.
For example, in an article published this week in GENETICS as part of the GSA journals’ new Multiparental Populations Collection, Tsaih et al. describe how they used HS rats to rapidly track down a gene associated with diabetic traits in humans.
In previous work, the group, led by Leah Solberg Woods (Medical College of Wisconsin), used QTL mapping in the HS rats to dissect a broad, 67 Mb chromosomal region that had been linked to glucose intolerance in rats, as well as to type 2 diabetes in humans. Within the region of interest, they found a 3.1 Mb QTL for glucose tolerance and insulin resistance. That was a massive improvement over the results from previous studies, but still left the team with 86 candidates genes.
In the new work, the researchers narrowed down their list by looking for genes that were differentially expressed between glucose intolerant rats and those with normal glucose regulation. One of these genes, Tpcn2, was located within the 3.1 Mb QTL. Tpcn2 turned up again when the authors tested 18 SNPs across this region for association with fasting glucose levels. The only two variants with significant associations both mapped to the Tpcn2 region, and they also affected Tpcn2 expression. The team then used mice to test the effect of knocking out the homolog of Tpcn2. Consistent with a role for the gene in glucose regulation, the knock-out mice had lower fasting glucose levels than wild-type.
Tpcn2 encodes Two-Pore Channel 2 (TPC2), an ion channel suggested to be a receptor for the calcium-mobilizing agent NAADP, which may play a role in glucose homeostasis. Others argue TPC2 forms a complex with mTOR that regulates lysosomal function in response to nutrient status. Either way, it seems plausible that the gene might affect glucose regulation.

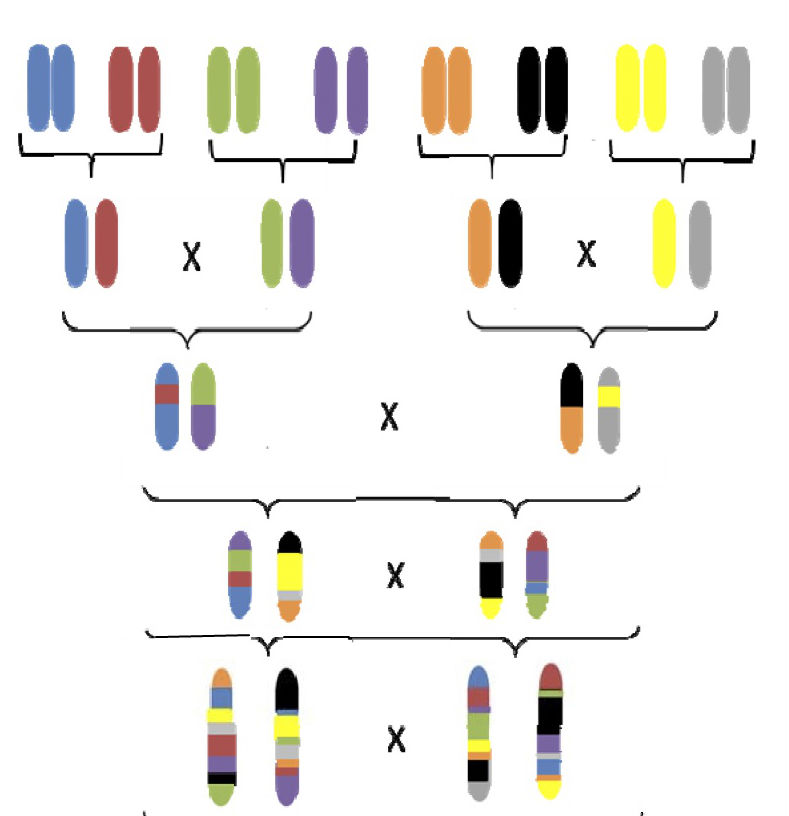
In the Heterogeneous Stocks breeding scheme, eight genetically distinct inbred founders are intercrossed to generate a balanced distribution of recombined alleles in their descendants. Image credit: Katie Holl and Leah Solberg Woods.
All these data make a good case that Tpcn2 can affect diabetic traits in rodents, but what about humans? The authors turned for clues to two large human genetics consortia: Diabetes Genetics Replication and Meta-analysis (DIAGRAM) and Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC). Of the 14 common em>Tpcn2 SNPs represented within these datasets, none were associated with fasting glucose or diabetes, but two did show significant associations with fasting insulin, another key diabetic trait intimately tied to glucose levels.
So, in only a few years, with the help of the HS rats, the authors were able to map a medically-relevant complex trait in rats, and then identify a plausible causal gene that may have related effects in humans. Other groups are also using the HS rats to map QTLs, with results published for traits like arterial defects, bone mineral density, T-cell selection, and fear behavior. Such valuable work owes a debt to those who, in times of economic hardship, developed and defended a long-term community resource.
To explore more work in the GSA Journals’ Multiparental Populations Collection, visit the collection home page.
MPP Collection press release: Want to link genes to complex traits? Start with more diversity.
Citation:
Tsaih S.W., S. Jia, M. Kaldunski, M. Tschannen, H. He, J. W. Andrae, S.-H. Li, A. Stoddard, A. Wiederhold & J. Parrington & (2014). Identification of a Novel Gene for Diabetic Traits in Rats, Mice, and Humans, GENETICS, 198 (1) 17-29. DOI: http://dx.doi.org/10.1534/genetics.114.162982