Multiparental populations (MPPs) have brought a new era in mapping complex traits, as well as new analytical challenges. To face these challenges and encourage innovation, the GSA journals launched the ongoing Multiparental Populations series in 2014. This month’s issues of GENETICS and G3 feature a bumper 16 MPP articles, timed to celebrate a new easy-to-use site for browsing the series. In line with our goal of encouraging communication across disciplinary boundaries, the “MPP People” profiles aim to introduce series authors working in a wide range of systems.
Not all sand field crickets can fly. The “short wing” morph of this species is grounded by its stumpy wings and feeble flight muscles. But because female short-wings don’t need to sink their limited resources into the costly trappings of flight, they are champion reproducers. In contrast, the “long wing” morphs can fly to new and better habitats, but produce substantially fewer eggs than their Earth-bound peers.
Libby King has long been fascinated by such dramatic tradeoffs in how organisms allocate their limited pool of energy to key traits, particularly in how they coordinate their allocation strategy with the nutritional environment.
“If they have a lot of resources and a big pool of energy, then there might be one optimal way to divvy up that pie, but if they’re really resource limited, then there might be another, better way,” says King.
During her PhD research with Daphne Fairbairn and Derek Roff at the University of California, Riverside, King investigated how the proportion of long-wing to short-wing morphs in a cricket population is influenced by variation in resource availability across the landscape. From this ecological and evolutionary perspective, King was drawn to thinking about the mechanisms behind these patterns. What are the genes involved in strategy variation? There were no easy answers.
“That’s how I got pulled into the very hard problem of how to dissect a very complex phenotype,” says King. To work on such problems, she shifted focus in her postdoc research to fruit fly genomics.

Elizabeth G. King, University of Missouri–Columbia
“Libby is really talented and has already made big contributions to the field,” says her former postdoc mentor Anthony Long at the University of California, Irvine. “She is one of the rare people with a really deep appreciation for both classical quantitative genetics and more modern molecular approaches to the dissection of complex traits.”
Working with Long and Stuart Macdonald (University of Kansas), she played a key role in the development and testing of the Drosophila Synthetic Population Resource (DSPR), a pair of multiparental populations with high power and resolution for complex trait mapping.
The DSPR is derived from 15 founder inbred lines capturing fruit fly genetic diversity from around the world. Each of the two replicate DSPR populations was created by crossing eight founder lines (seven unique, one shared by both replicates) for 50 generations and then establishing more than 800 recombinant inbred lines (RILs) per population.
Some of the main advantages of the DSPR, says King, include the very high mapping resolution and the fact that initial trait mapping requires only phenotyping of the RILs because the genome sequence of each can be imputed. During her postdoc, King developed a Hidden Markov Model method to infer the RIL sequences using dense genotyping with RAD markers and the known genome sequences of the founders.
Inbred lines also enable testing multiple phenotypes across the same genotypes. This last feature is particularly important for King now that she has established her own lab and is using the DSPR to investigate resource allocation in Drosophila.
“These are pretty complex traits that encompass all sorts of other traits, so it’s really useful to be able to measure phenotypes at those multiple levels of organization,” she says.
In a paper published in the MPP series in the June issue of GENETICS, King’s group, led by graduate student Patrick Stanley, examined a widespread resource allocation pattern: in many eukaryotes, individuals tend to live longer and reproduce less when nutrients are scarce. This is hypothesized to help conserve resources for survival while the individual waits for conditions to improve.
One potential avenue for the evolution of this pattern is the highly conserved insulin/insulin-like growth factor/Target of Rapamycin (IIS/TOR) pathway. In many models, knockouts for genes in this pathway live longer than wild-type, and there is substantial evidence that the pathway is involved in coordinating growth and metabolism with nutritional conditions. Changing expression of these IIS/TOR genes is often hypothesized to drive the lifespan/reproduction shift seen in low nutrient conditions.
Stanley et al. used the DSPR to investigate the link between this pathway and the nutrient-induced lifespan shift by focusing on 56 core IIS/TOR genes. They assayed how expression of these genes changed on three different diets, mapped genetic variation influencing these responses, and then measured lifespan of a subset of flies under these conditions.
As expected, most genes in the pathway changed expression between diet conditions. The team successfully mapped the genetic basis for these changes, including two trans QTLs that likely represent transcription factors that respond to diet.
But, consistent with mixed results emerging elsewhere in the literature, the results do not provide strong evidence that IIS/TOR gene expression changes drive the lifespan response to diet. They observed relatively small expression changes in most genes, rather than strong changes at a few key genes, and these were not in the direction you would predict based on knockout mutant phenotypes. “There is a change in global gene expression in response to diet, and this may well be partially driving what’s going on, but it’s clearly not the whole story,” says King.
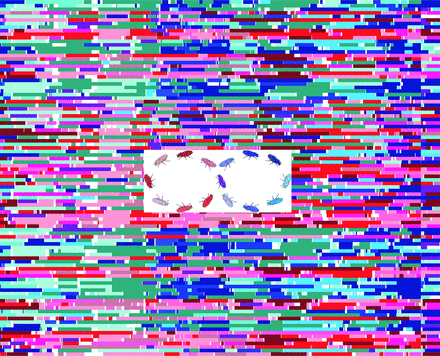
Detail from the cover of the July 2012 issue of GENETICS, which included an article describing the properties of the Drosophila Synthetic Population Resource (DSPR). The center image represents the 15 inbred founder lines used to create the DSPR (blue = A population, red = B population, center purple fly representing the founder line shared between the populations). The background shows a sample of 100 RILs with the colors representing the founder ancestry across the genome. White areas represent an uncertain founder assignment.
A second MPP paper by King and Long in the June issue of G3 uses simulation to explore how an important bias affecting QTL mapping applies to large panels like the DSPR. This bias, known as the Beavis effect, is the tendency of significant QTLs to have overestimated effect sizes. This is because a test with an overestimated effect size is more likely to be found significant than one with an underestimated effect.
This phenomenon is one reason that, even for a true hit, validating a significant locus in a second population can fail if the experimental design lacks the power to detect the QTL at its true effect size. Although the Beavis effect is well established for traditional two-way QTL mapping, its importance is unknown for multiparental designs and association mapping, which can involve millions of tests.
The study revealed that the most important factor affecting the strength of the Beavis effect is the sample size, with the detail of the mapping design mattering much less. In essence, the more lines that are phenotyped, the weaker the Beavis effect becomes and the more accurate the QTL effects estimates become. Using only a few lines from the DSPR would make it is highly likely that the effect sizes of significant QTL are overestimated, and they may be difficult to validate in replicate experiments or cross-validate between different mapping designs. King and Long use their results to provide guidelines on the sample sizes needed to accurately estimate the percent effect variance of an identified QTL and the conditions under which a mapped QTL is likely to be successfully replicated.
King’s work with the DSPR has not only provided her the means to pursue her interest in the evolution of resource allocation strategies. It has also enriched the field as a whole, providing a powerful new component to the Drosophila toolkit for unraveling the molecular mechanisms of complex traits.
Read other MPP People profiles.
Browse the GSA Journals MPP series.
MPP AUTHOR:
Elizabeth G. King, University of Missouri–Columbia
MPP ARTICLES:
The Beavis Effect in Next-Generation Mapping Panels in Drosophila melanogaster
Elizabeth G. King and Anthony D. Long
Patrick D. Stanley, Enoch Ng’oma, Siri O’Day, and Elizabeth G. King
The genetic architecture of methotrexate toxicity is similar in Drosophila melanogaster and humans
, , , and
, , and
Using Drosophila melanogaster to identify chemotherapy toxicity genes
, , and
, and













