Check out the April issue of GENETICS by looking at the highlights or the full table of contents!
ISSUE HIGHLIGHTS
Mismatch repair incompatibilities in diverse yeast populations, pp. 1459-1471
Duyen T. Bui, Anne Friedrich, Najla Al-Sweel, Gianni Liti, Joseph Schacherer, Charles F Aquadro, and Eric Alani
Microbial populations with elevated mutation rates can adapt more rapidly to new environments. Bui et al. hypothesized that a naturally occurring DNA mismatch repair gene incompatibility could yield mutator progeny that can rapidly but transiently adapt to environmental stress. They surveyed 1,010 natural yeast isolates for signatures of the incompatibility. Consistent with their hypothesis, an isolate with the incompatible genotype displayed a mutation rate similar to compatible strains due to the presence of extragenic suppressors, and isolates were identified that yield incompatible genotypes when sporulated.
The predicted cross value for genetic introgression of multiple alleles, pp. 1409-1423
Ye Han, John N. Cameron, Lizhi Wang, and William D. Beavis
Han et al. introduce a new metric, the Predicted Cross Value (PCV), for selecting breeding parents. Unlike estimated breeding values, which represent predictions of general combining ability, the PCV predicts specific combining ability The PCV takes estimates of recombination frequencies as an input vector and calculates the probability that a pair of parents will produce a gamete with desirable alleles at all specified loci.
Genomic rearrangements in Arabidopsis considered as quantitative traits, pp. 1425-1441
Martha Imprialou, Andre Kahles, Joshua G. Steffen, Edward J. Osborne, Xiangchao Gan, Janne Lempe, Amarjit Bhomra, Eric Belfield, Anne Visscher, Robert Greenhalgh, Nicholas P. Harberd, Richard Goram, Jotun Hein, Alexandre Robert-Seilaniantz, Jonathan Jones, Oliver Stegle, Paula Kover, Miltos Tsiantis, Magnus Nordborg, Gunnar Ratsch, Richard M. Clark, and Richard Mott
Structural Rearrangements can have unexpected effects on quantitative phenotypes. Surprisingly, these rearrangements can also be considered as quantitative phenotypes in their own right, in which QTLs contain genetic variation that correlates with presence of a structural variant, possibly elsewhere in the genome. Applying this idea to Arabidopsis, this study maps thousands of structural variants, about 25% of which are transpositions. It further shows they can explain more heritability of certain complex traits than standard genetic variation does.
A Bayesian approach for analysis of whole-genome bisulphite sequencing data identifies disease-associated changes in DNA methylation, pp. 1443-1458
Owen J. L. Rackham,Sarah R. Langley, Thomas Oates, Eleni Vradi, Nathan Harmston, Prashant K. Srivastava, Jacques Behmoaras, Petros Dellaportas, Leonardo Bottolo, and Enrico Petretto
Whole-genome bisulphite sequencing (WGBS) can identify important methylation differences between diseased and healthy samples. However, results from existing techniques are often strongly affected by changes in the underlying parameters. Rackham et al. developed a parameter-free approach (called ABBA) that outperforms existing methods across a variety of experimentally relevant settings.
Mobile introns shape the genetic diversity of their host genes, pp. 1641-1648
Jelena Repar and Tobias Warnecke
Self-splicing introns populate several highly conserved protein-coding genes in fungal and plant mitochondria. In fungi, many of these introns have retained their ability to spread to intron-free target sites, often assisted by intron-encoded endonucleases that initiate the homing process. Repar and Warnecke show for different fungal species that genetic diversity in exons increases as one approaches a mobile intron. They rule out relaxed purifying selection as the cause of uneven nucleotide diversity Instead, their findings implicate intron mobility as a direct driver of host gene diversity
Second-generation Drosophila chemical tags: sensitivity, versatility and speed, pp. 1399-1408
Ben Sutcliffe, Julian Ng, Thomas O. Auer, Mathias Pasche, Richard Benton, Gregory S. X. E. Jefferis, and Sebastian Cachero
Thick tissue specimens present major challenges for labeling cells and subcellular structures in a rapid and reliable manner. Sutcliffe et al. present improved second generation CLIP, SNAP, and Halo chemical labeling reagents. These multimerized tags are up to 64 times brighter than the first generation tools, while retaining their 100-fold increase in labeling speed compared to immunohistochemistry. They also developed LexAop2 reporters and conditional tools, including chemical Brainbow, that are activated by the orthogonal Bxb1 recombinase.
Wheat landrace genome diversity, pp. 1657-1676
Luzie U. Wingen, Claire West, Michelle Leverington-Waite, Sarah Collier, Simon Orford, Richard Goram, Cai-Yun Yang, Julie King, Alexandra M. Allen, Amanda Burridge, Keith J. Edwards, and Simon Griffiths
Understanding the genomic complexity of bread wheat is important for unraveling domestication processes, environmental adaptation, and for future improvement of the crop. A panel of 60 segregating bi-parental populations were developed between mainly landrace accessions and a modern elite reference parent. Genetic maps and a consensus map, containing 2,498 loci, were constructed. These newly developed tools were used to investigate the rules underlying genome fluidity of the hexaploid genome. Marker order was, with exceptions, highly correlated between maps. Interestingly, an astonishing number of genome translocations, many cases of segregation distortion, and 114 QTL involved in recombination rate were discovered.
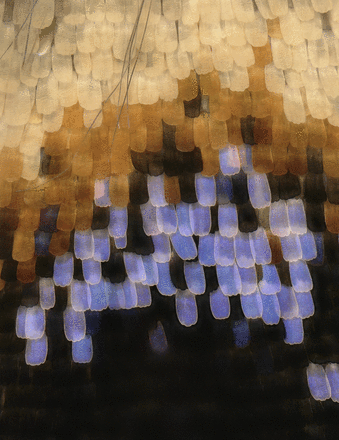
Genetic basis of melanin pigmentation in butterfly wings, pp. 1537-1550
Linlin Zhang, Arnaud Martin, Michael W. Perry, Karin R. L. van der Burg, Yuji Matsuoka, Antonia Monteiro, and Robert D. Reed
Butterfly wing patterns are a model system for studying the evolution and development of adaptive traits. Zhang et al. combine RNA-seq and CRISPR/Cas9 genome editing to characterize and functionally assess genes involved in butterfly wing pattern development. Focusing their approach on the melanin pigmentation pathway showed that while some melanin genes have deeply conserved function across insects, some play expanded roles in butterfly wings to tune coloration of specific pattern elements.